HLA typing by sequence-specific primers (PCR-SSP)
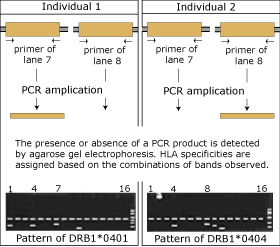 |
| Amplification with sequence-specific primers yields only a product if the target sequences are present in the DNA sample (compare lane 7 and 8 with the figure) In total 16 primers are used for the analysis of HLA-DR4 allele. |
|
The techniques
The procedure relies on the specificity of primer extension that is matched
or mismatched with the template at its 3’end. A combination of 2
primers designed for each of 2 polymorphic sequence motifs in cis allows
the identification of an allele or a group of alleles that are chracterized
by these 2 motifs. The presence or absence of the 2 motifs in cis is usually
detected by gel electrophoresis, but other detection methods have been
developped. The method is rapid and ideally suited for small numbers of
samples. Because very few sequences are absolutely allele specific, SSP combine
several primer to discriminate a particular allele unambigously.
Resolution
The method can be applied for low resolution typing, using primer combinations
that detect all alleles within a given serotype, as well as for high resolution
typing. Low resolution HLA-A/B/DR typing can be achieved by a combination
of 72 primer mixes. Depending on the HLA phenotype, a combination of 300-400
different primer mixes are necessary to determine the HLA-A/B/C/DRB1/DRB3/DRB4/DRB5/DQB1
alleles in a given individual.
|


 Print
Print